generate_cells() runs simulations in order to determine the gold standard
of the simulations.
simulation_default() is used to configure parameters pertaining this process.
generate_cells(model)
simulation_default(
burn_time = NULL,
total_time = NULL,
ssa_algorithm = ssa_etl(tau = 30/3600),
census_interval = 4,
experiment_params = bind_rows(simulation_type_wild_type(num_simulations = 32),
simulation_type_knockdown(num_simulations = 0)),
store_reaction_firings = FALSE,
store_reaction_propensities = FALSE,
compute_cellwise_grn = FALSE,
compute_dimred = TRUE,
compute_rna_velocity = FALSE,
kinetics_noise_function = kinetics_noise_simple(mean = 1, sd = 0.005)
)
simulation_type_wild_type(
num_simulations,
seed = sample.int(10 * num_simulations, num_simulations)
)
simulation_type_knockdown(
num_simulations,
timepoint = runif(num_simulations),
genes = "*",
num_genes = sample(1:5, num_simulations, replace = TRUE, prob = 0.25^(1:5)),
multiplier = runif(num_simulations, 0, 1),
seed = sample.int(10 * num_simulations, num_simulations)
)Arguments
- model
A dyngen intermediary model for which the gold standard been generated with
generate_gold_standard().- burn_time
The burn in time of the system, used to determine an initial state vector. If
NULL, the burn time will be inferred from the backbone.- total_time
The total simulation time of the system. If
NULL, the simulation time will be inferred from the backbone.- ssa_algorithm
Which SSA algorithm to use for simulating the cells with
GillespieSSA2::ssa()- census_interval
A granularity parameter for the outputted simulation.
- experiment_params
A tibble generated by rbinding multiple calls of
simulation_type_wild_type()andsimulation_type_knockdown().- store_reaction_firings
Whether or not to store the number of reaction firings.
- store_reaction_propensities
Whether or not to store the propensity values of the reactions.
- compute_cellwise_grn
Whether or not to compute the cellwise GRN activation values.
- compute_dimred
Whether to perform a dimensionality reduction after simulation.
- compute_rna_velocity
Whether or not to compute the propensity ratios after simulation.
- kinetics_noise_function
A function that will generate noise to the kinetics of each simulation. It takes the
feature_infoandfeature_networkas input parameters, modifies them, and returns them as a list. Seekinetics_noise_none()andkinetics_noise_simple().- num_simulations
The number of simulations to run.
- seed
A set of seeds for each of the simulations.
- timepoint
The relative time point of the knockdown
- genes
Which genes to sample from.
"*"for all genes.- num_genes
The number of genes to knockdown.
- multiplier
The strength of the knockdown. Use 0 for a full knockout, 0<x<1 for a knockdown, and >1 for an overexpression.
Value
A dyngen model.
See also
dyngen on how to run a complete dyngen simulation
Examples
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
model <-
initialise_model(
backbone = backbone_bifurcating(),
simulation = simulation_default(
ssa_algorithm = ssa_etl(tau = .1),
experiment_params = bind_rows(
simulation_type_wild_type(num_simulations = 4),
simulation_type_knockdown(num_simulations = 4)
)
)
)
# \donttest{
data("example_model")
model <- example_model %>% generate_cells()
#> Warning: Simulation does not contain all gold standard edges. This simulation likely suffers from bad kinetics; choose a different seed and rerun.
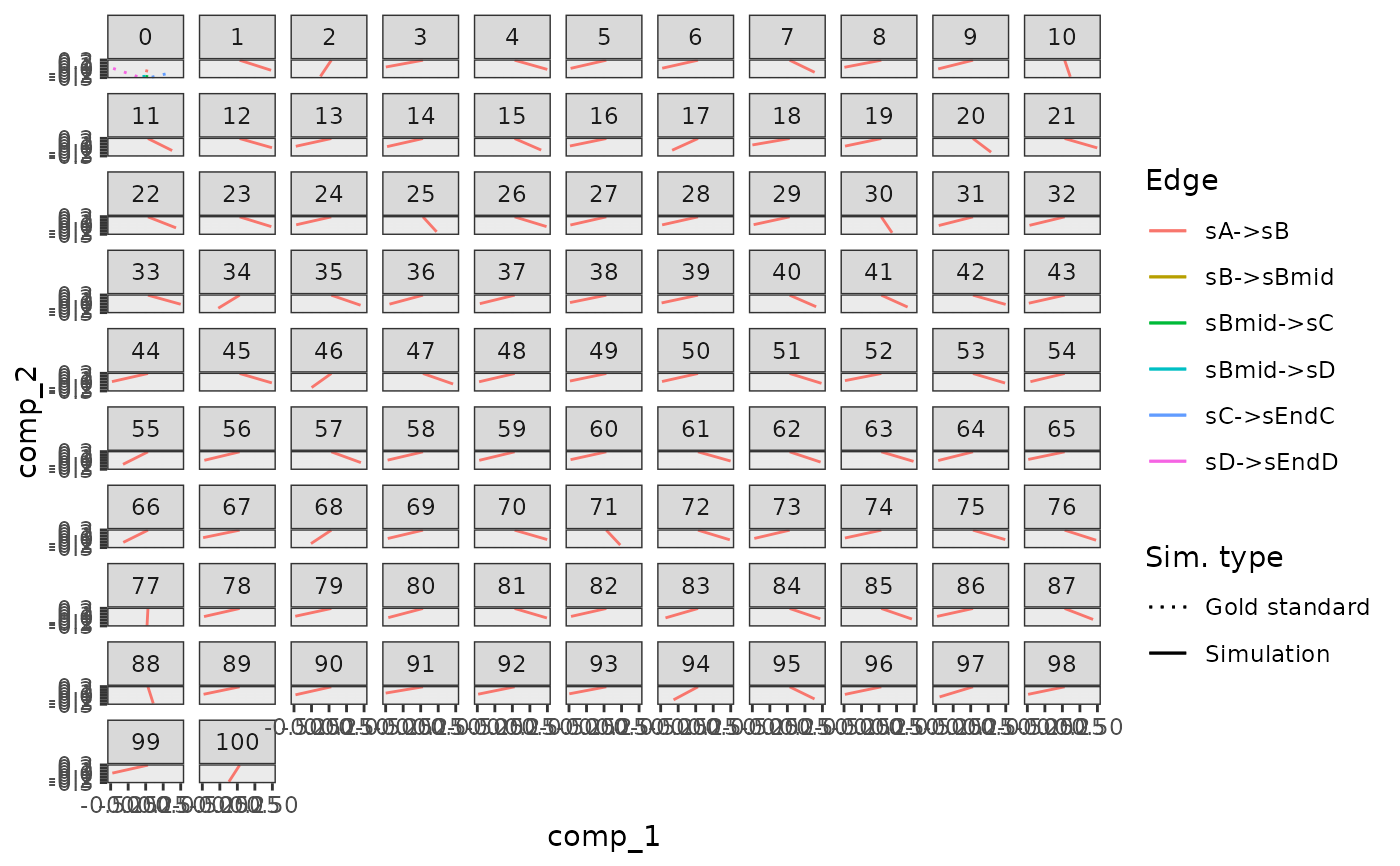
plot_simulations(model)
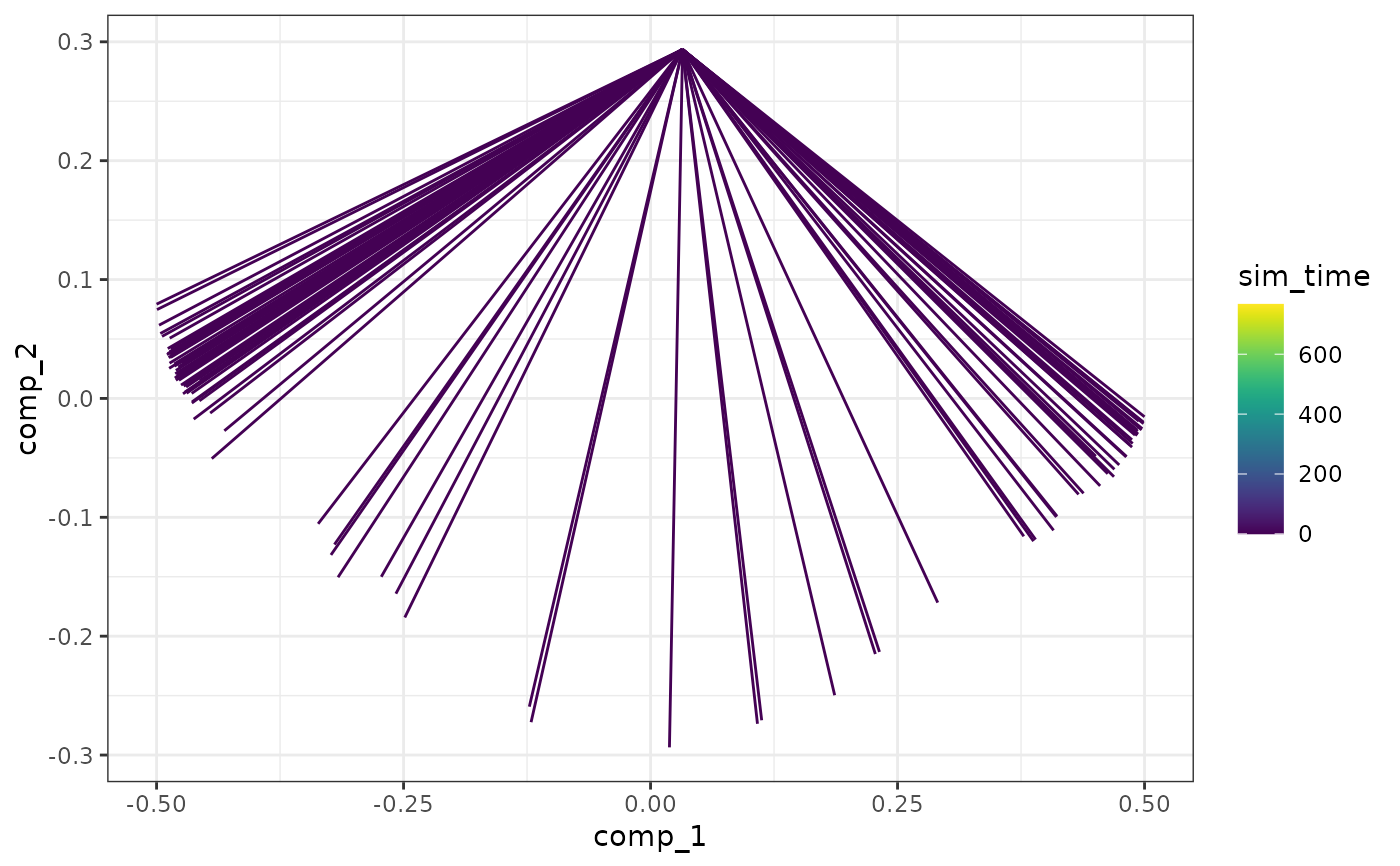 plot_gold_mappings(model)
plot_gold_mappings(model)
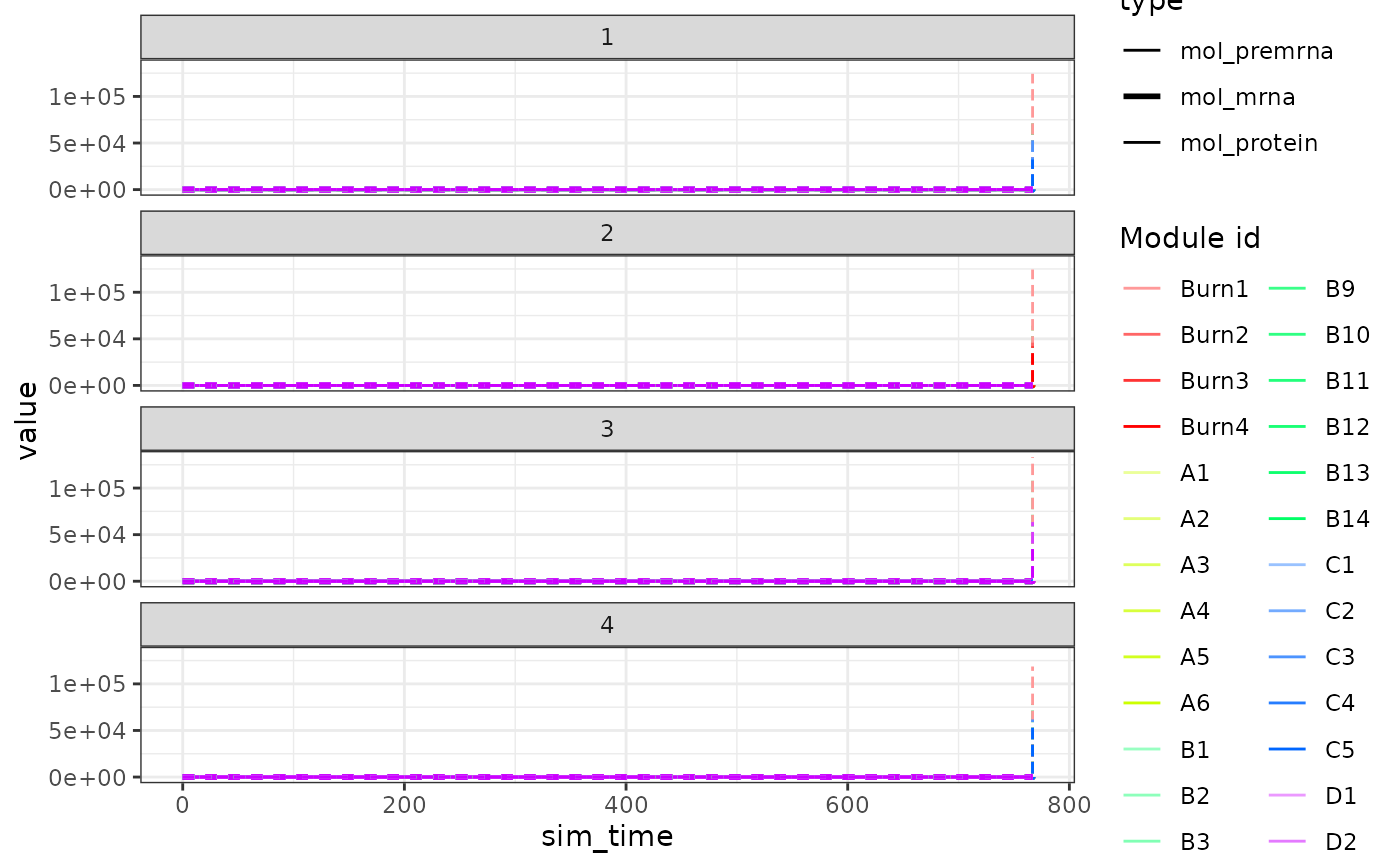
 plot_simulation_expression(model)
plot_simulation_expression(model)
 # }
# }