This vignette demonstrates the different dynamic processes topologies (e.g. bifurcating and cyclic). Note that, for the sake of reducing runtime for generating this vignette, the simulations are ran with reduced settings. Check out the vignette on tweaking parameters for information on how the different parameters are changed.
You can find a full list of backbones using ?list_backbones. This vignette will showcase each of them individually.
Linear
set.seed(1)
backbone <- backbone_linear()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot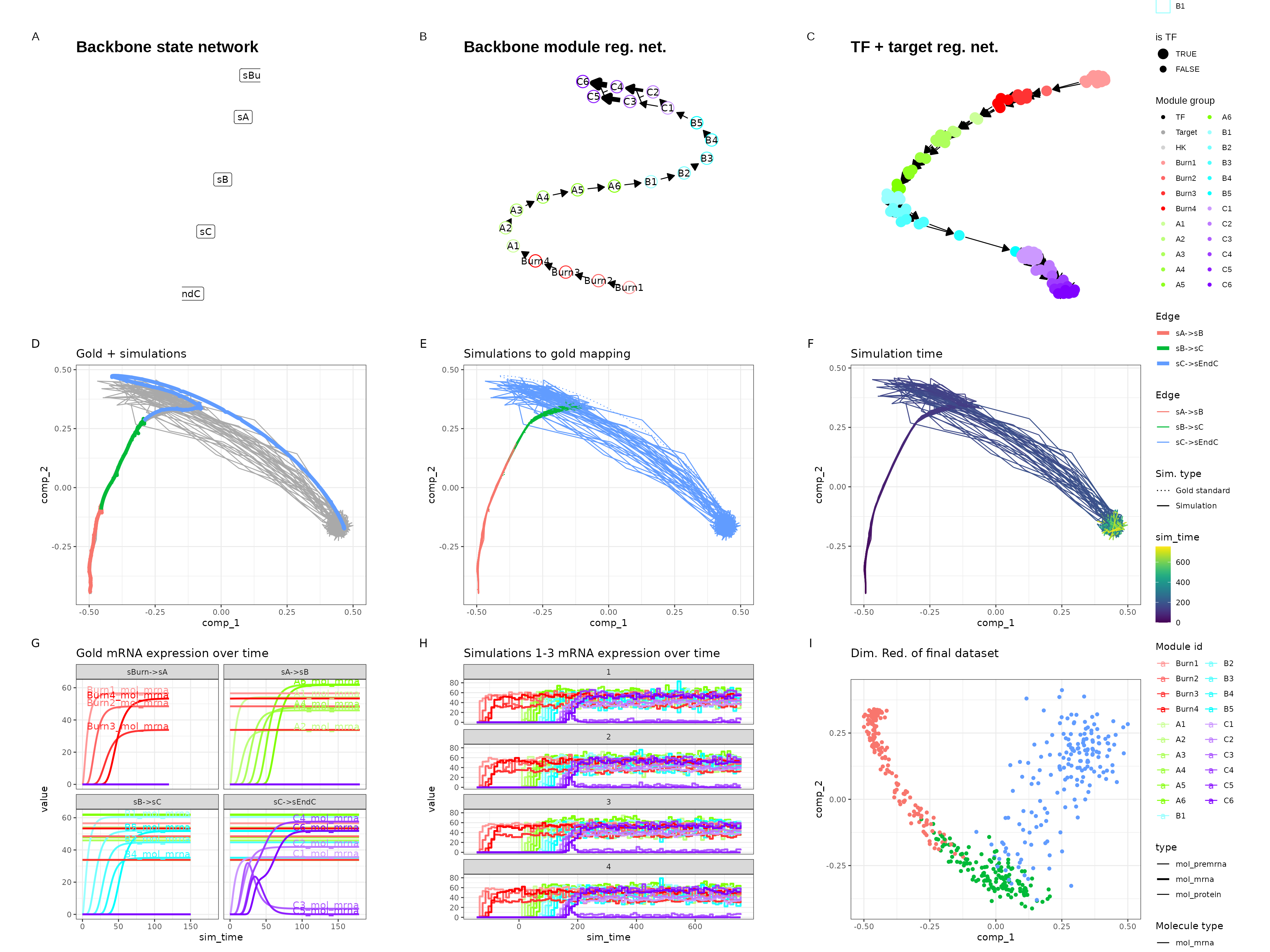
Bifurcating
set.seed(2)
backbone <- backbone_bifurcating()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot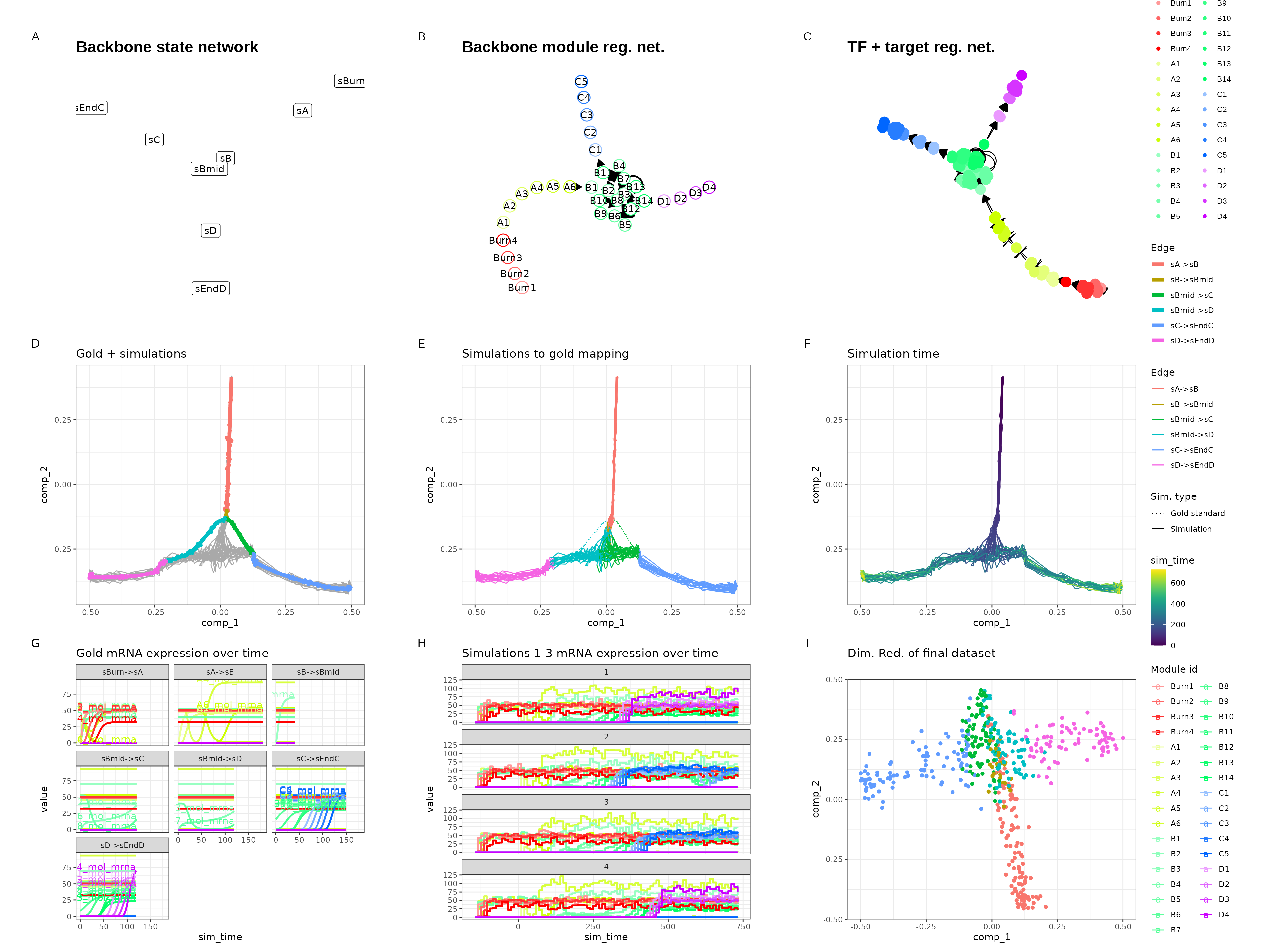
Bifurcating converging
set.seed(3)
backbone <- backbone_bifurcating_converging()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot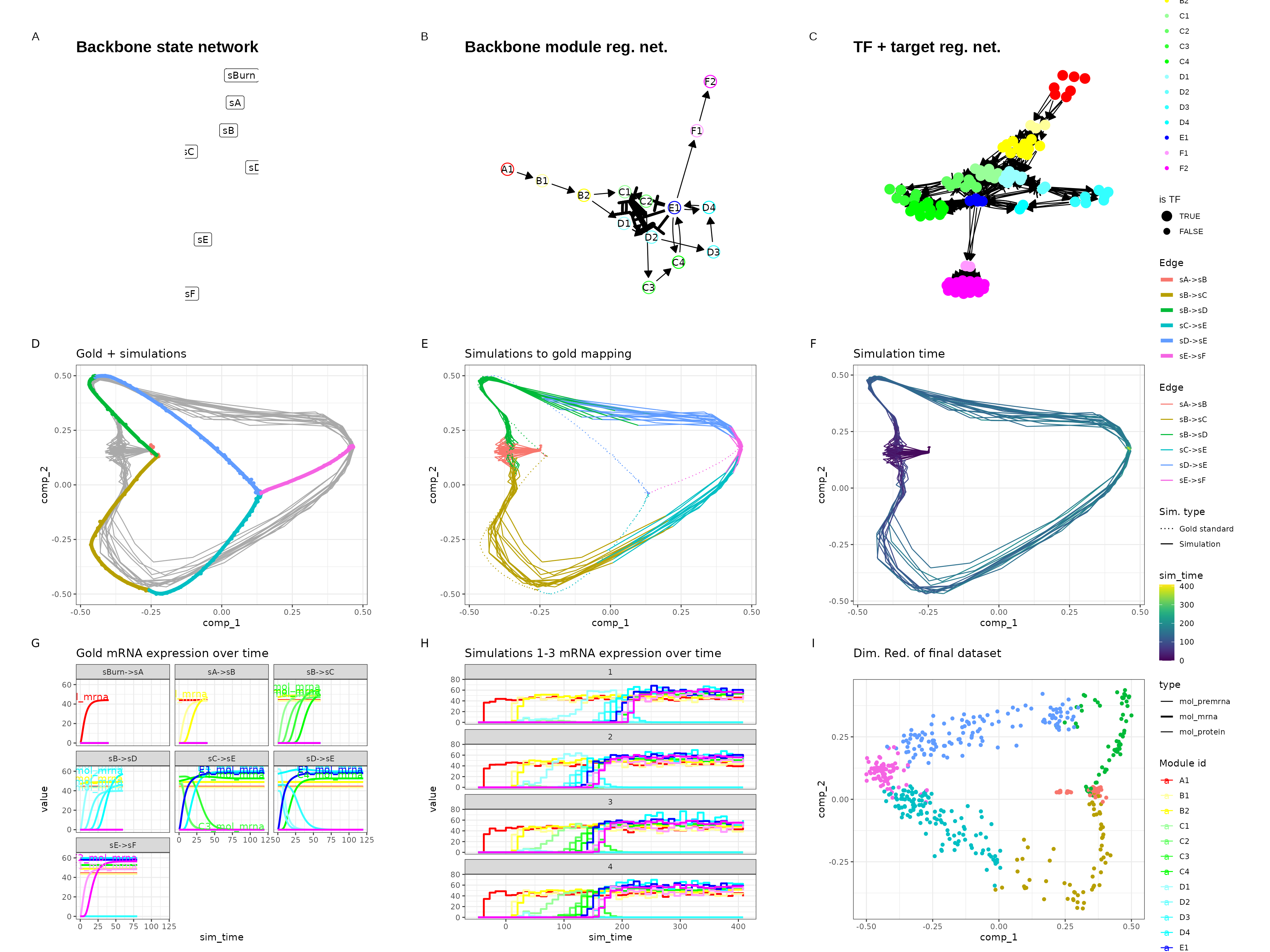
Bifurcating cycle
set.seed(4)
backbone <- backbone_bifurcating_cycle()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot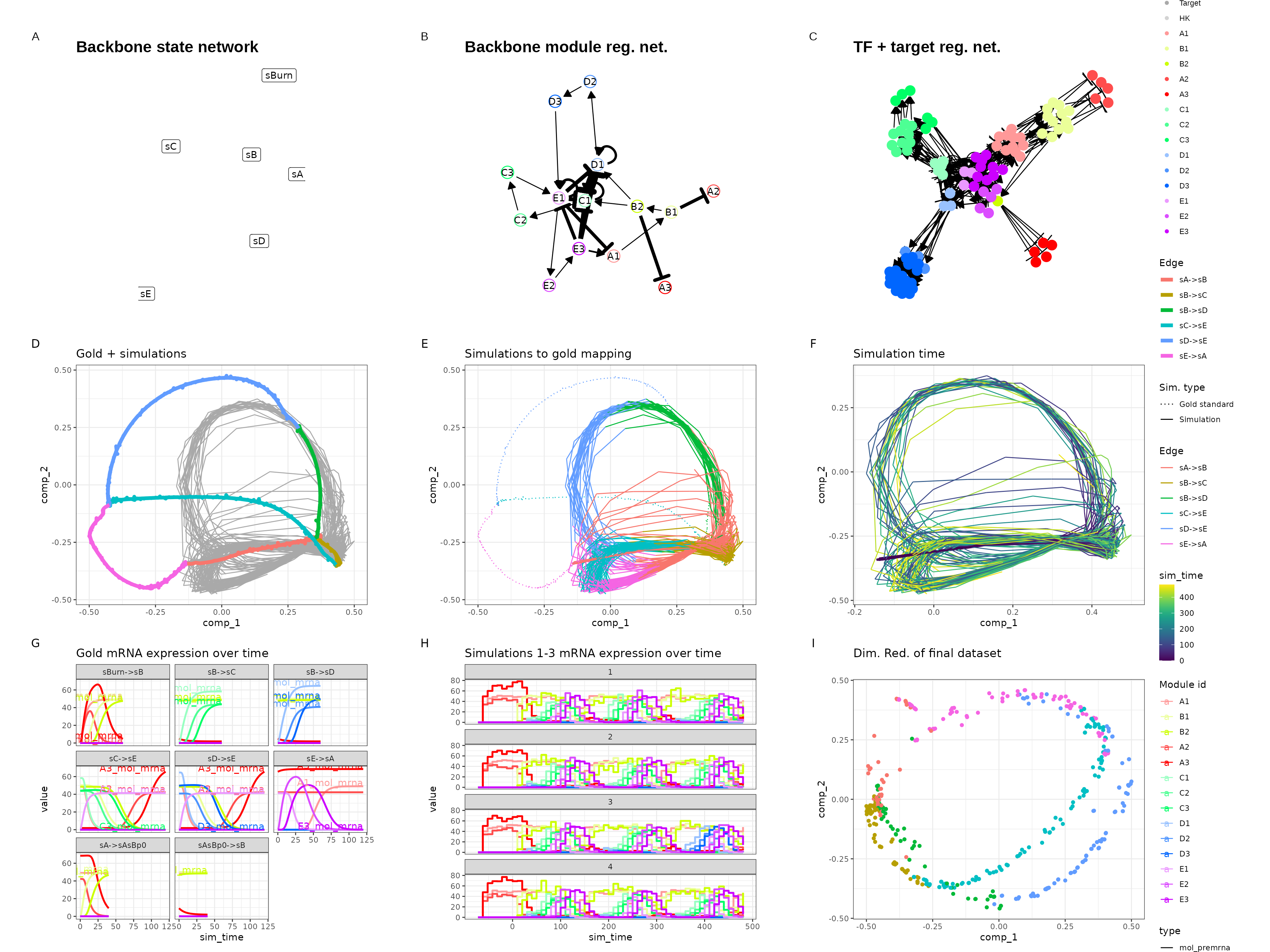
Bifurcating loop
set.seed(5)
backbone <- backbone_bifurcating_loop()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot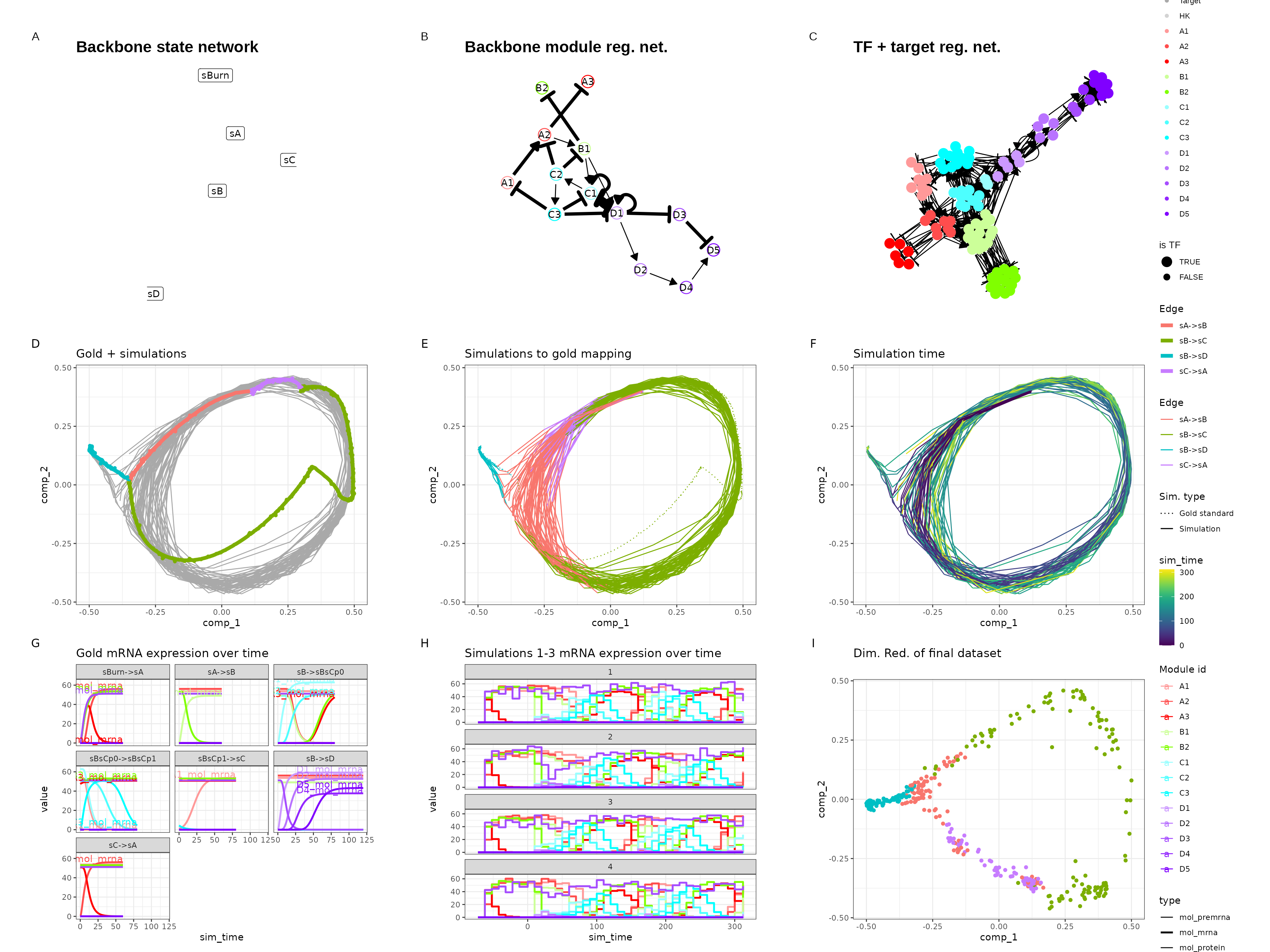
Binary tree
set.seed(6)
backbone <- backbone_binary_tree(
num_modifications = 2
)
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot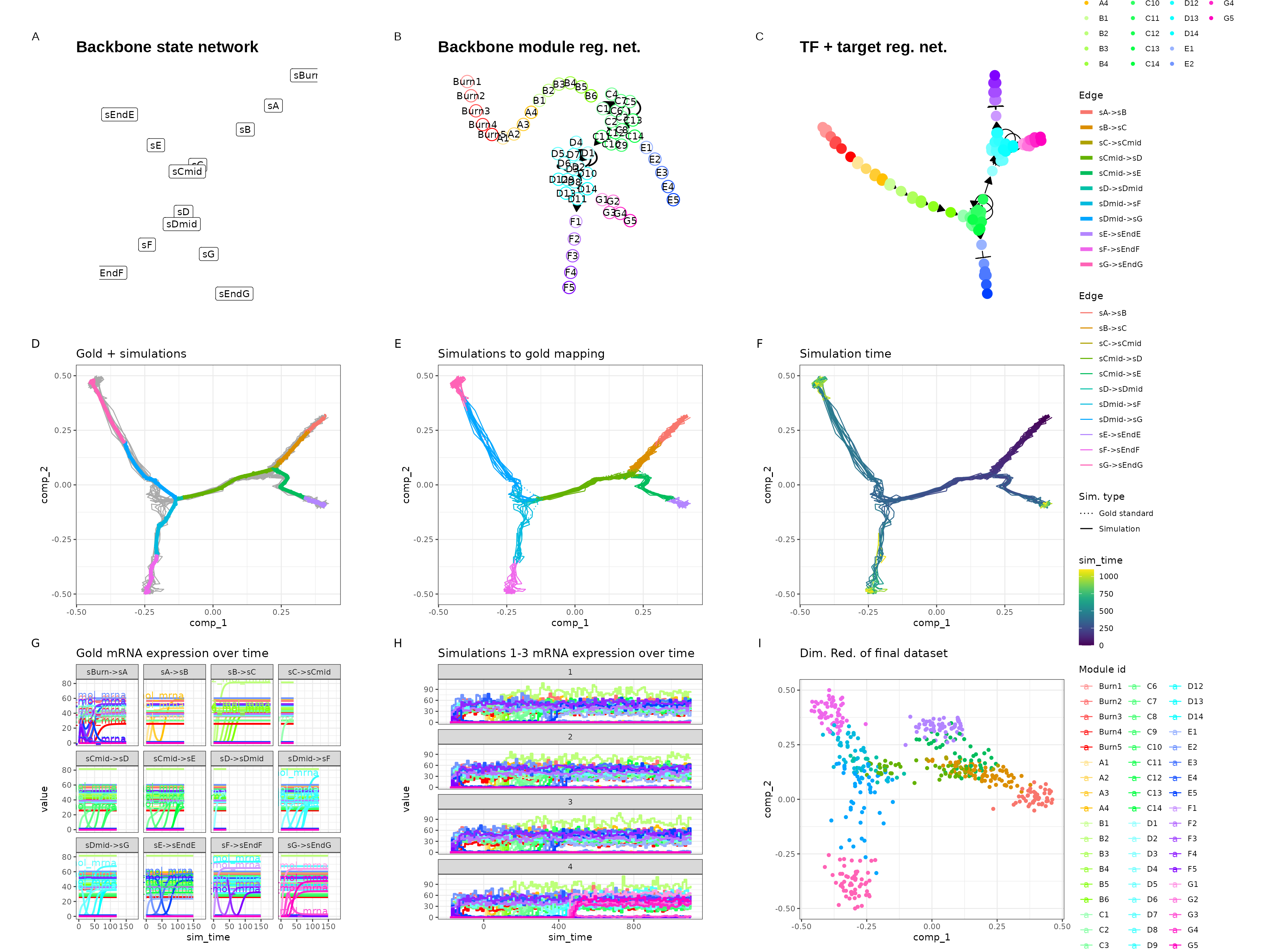
Branching
set.seed(7)
backbone <- backbone_branching(
num_modifications = 2,
min_degree = 3,
max_degree = 3
)
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in .generate_cells_predict_state(model): Simulation does not contain all
#> gold standard edges. This simulation likely suffers from bad kinetics; choose a
#> different seed and rerun.
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : Certain backbone segments are not covered by any of the simulations. If this is intentional, please ignore this warning.
#> Otherwise, increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot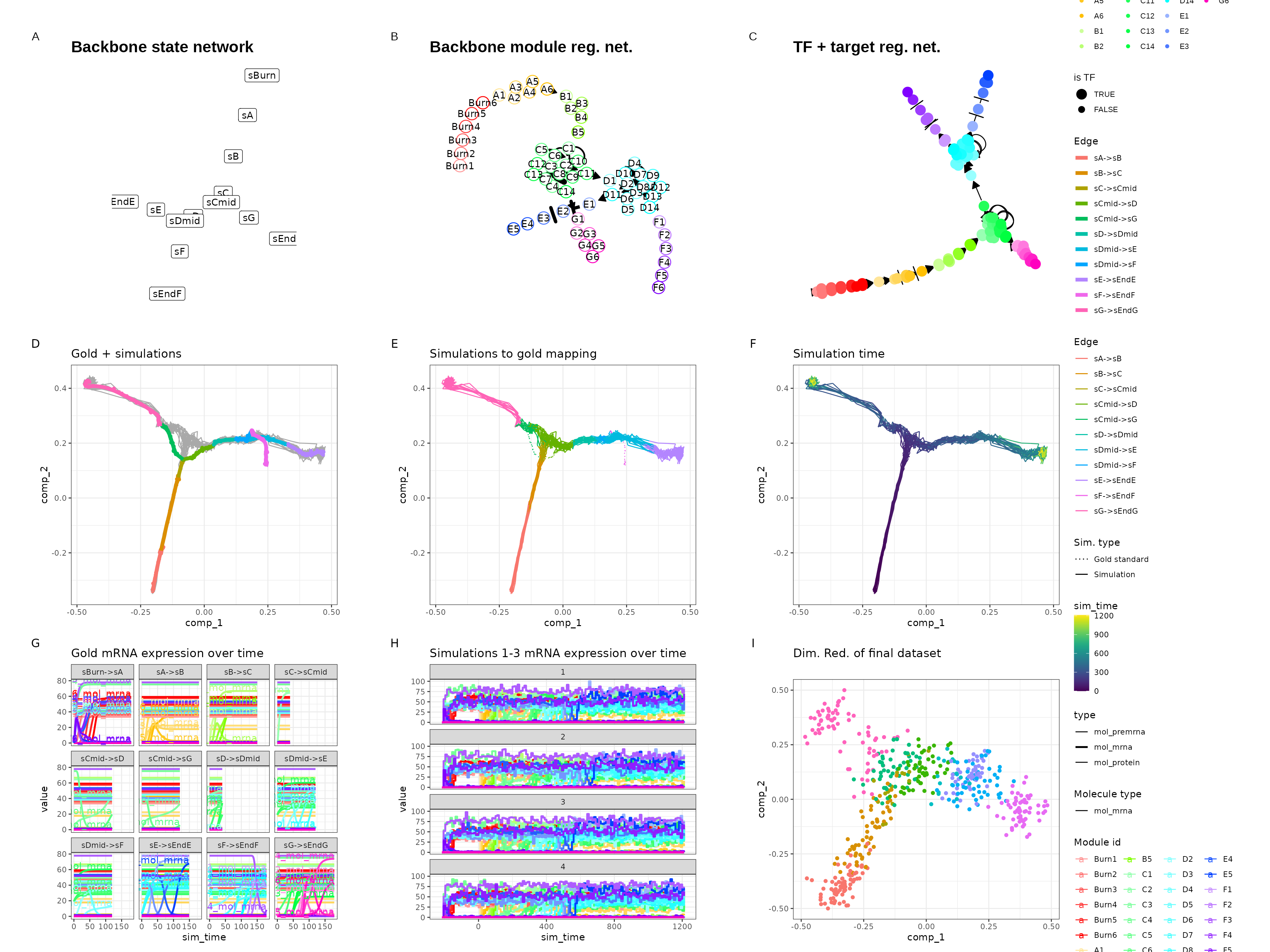
Consecutive bifurcating
set.seed(8)
backbone <- backbone_consecutive_bifurcating()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in .generate_cells_predict_state(model): Simulation does not contain all
#> gold standard edges. This simulation likely suffers from bad kinetics; choose a
#> different seed and rerun.
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : Certain backbone segments are not covered by any of the simulations. If this is intentional, please ignore this warning.
#> Otherwise, increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot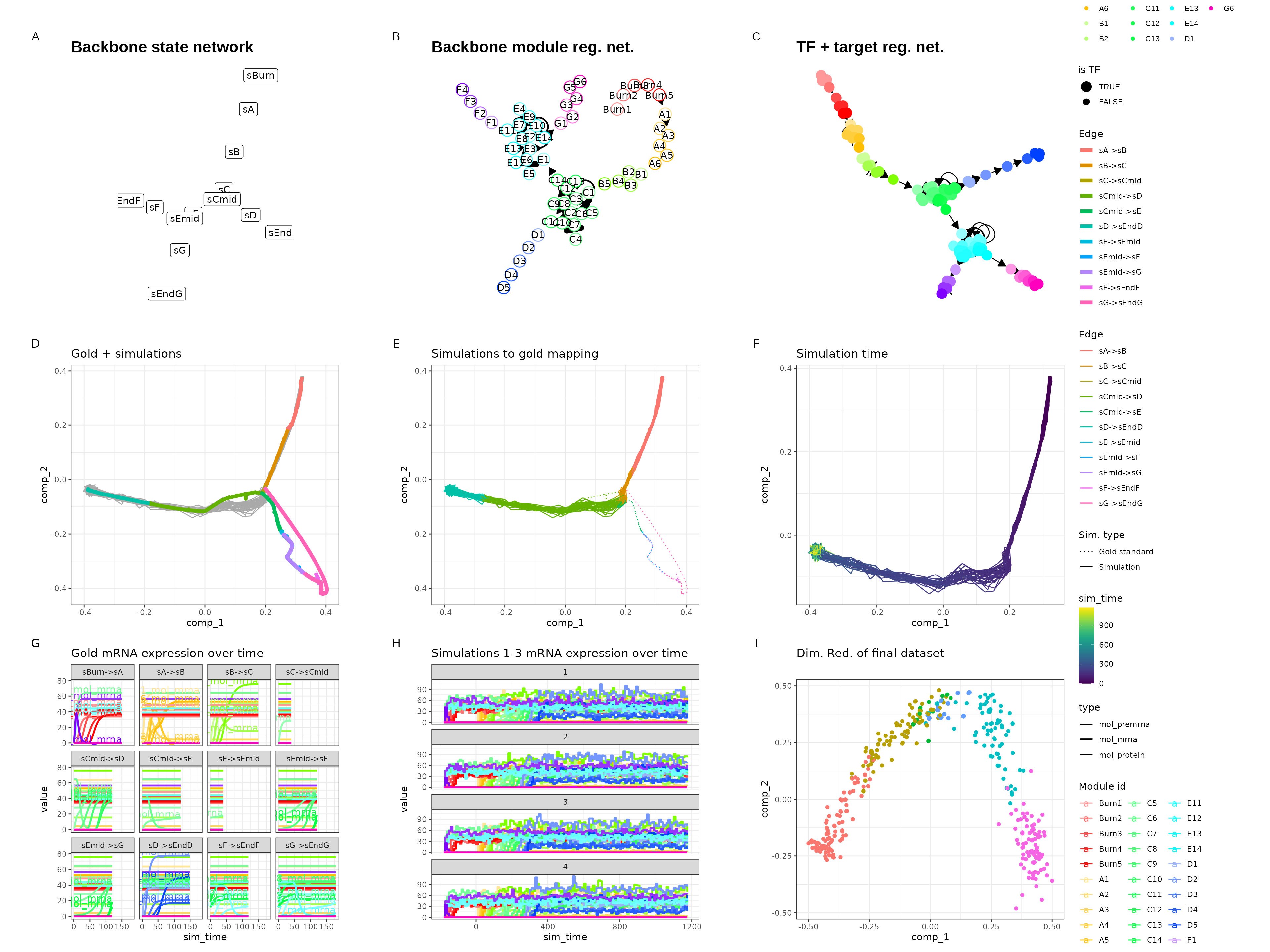
Trifurcating
set.seed(9)
backbone <- backbone_trifurcating()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot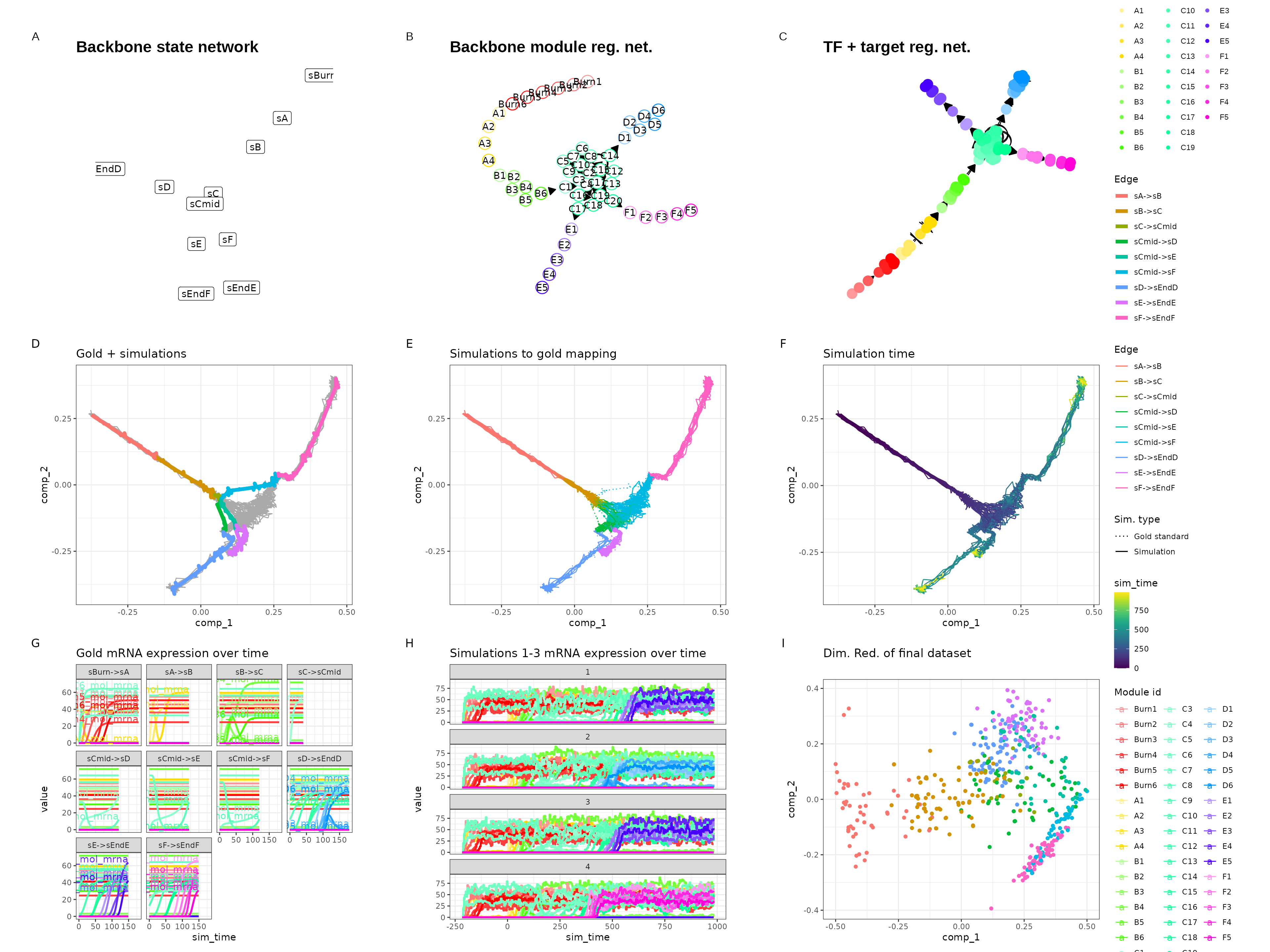
Converging
set.seed(10)
backbone <- backbone_converging()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot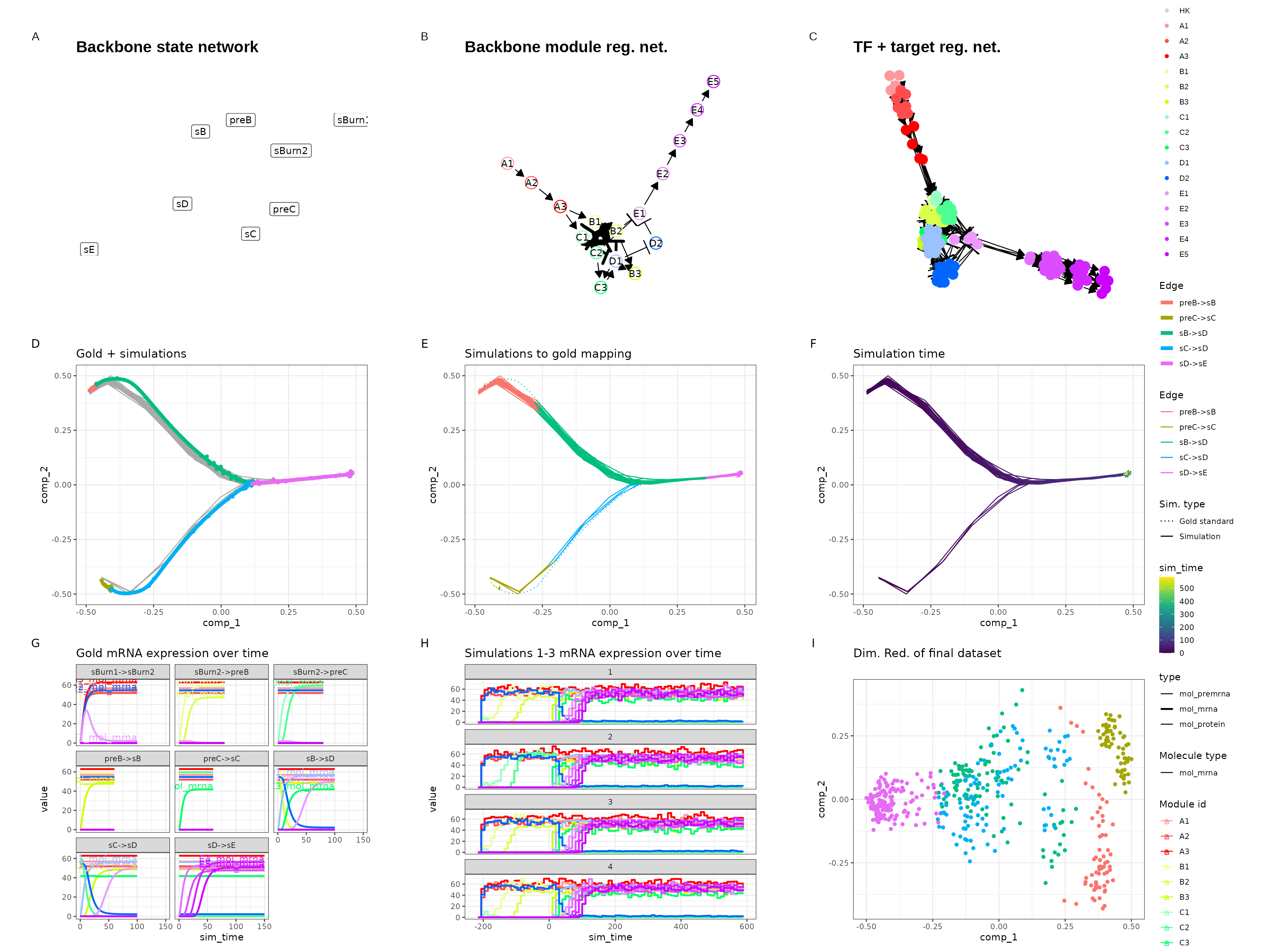
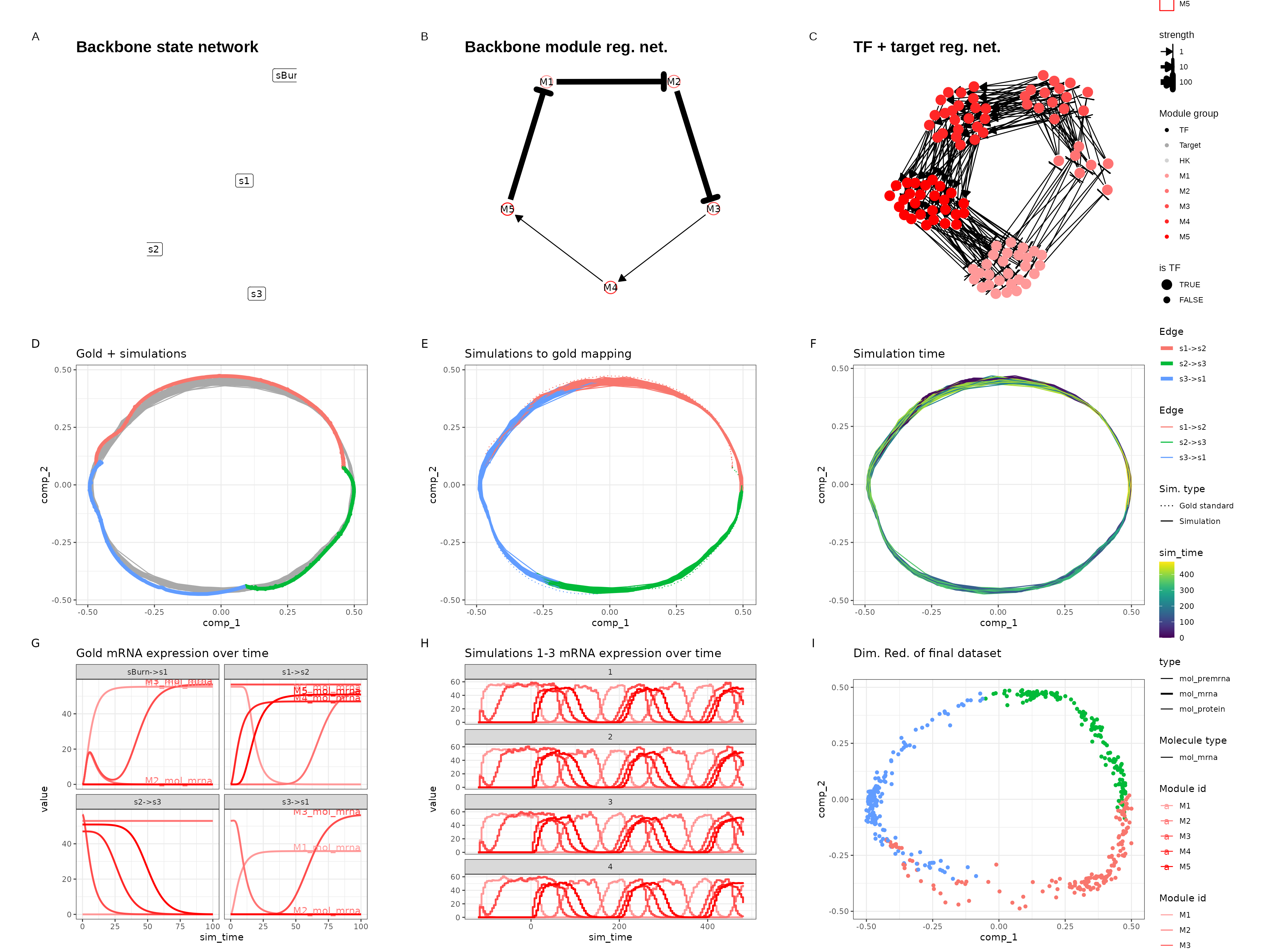
Cycle
set.seed(11)
backbone <- backbone_cycle()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot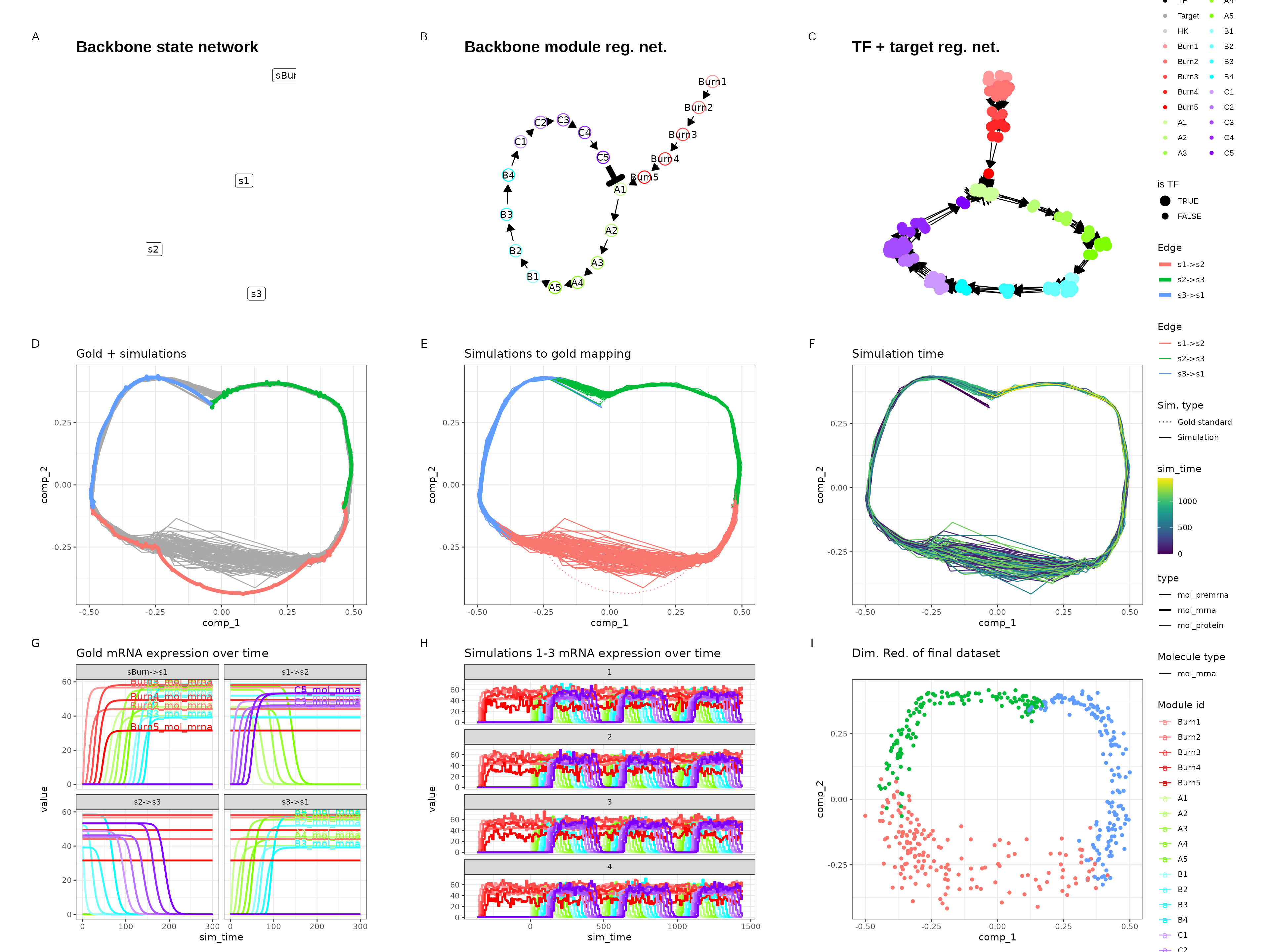
Disconnected
set.seed(12)
backbone <- backbone_disconnected()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
simulation_params = simulation_default(census_interval = 10, ssa_algorithm = ssa_etl(tau = 300 / 3600)),
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
#> Warning in .generate_cells_predict_state(model): Simulation does not contain all
#> gold standard edges. This simulation likely suffers from bad kinetics; choose a
#> different seed and rerun.
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : Certain backbone segments are not covered by any of the simulations. If this is intentional, please ignore this warning.
#> Otherwise, increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
#> Warning in params$fun(network = network, sim_meta = sim_meta, params = model$experiment_params, : One of the branches did not obtain enough simulation steps.
#> Increase the number of simulations (see `?simulation_default`) or decreasing the census interval (see `?simulation_default`).
out$plot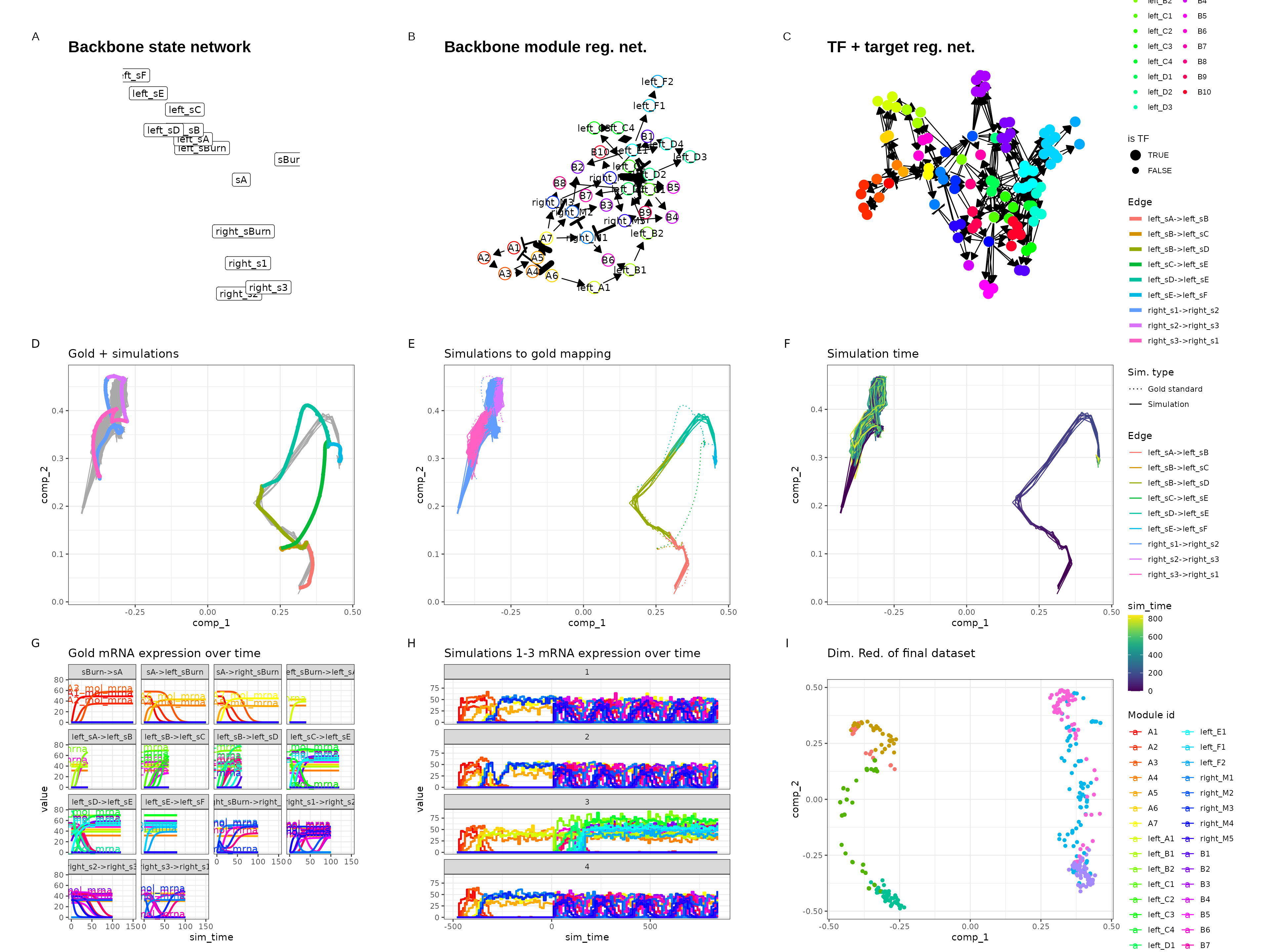
Linear simple
set.seed(13)
backbone <- backbone_linear_simple()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot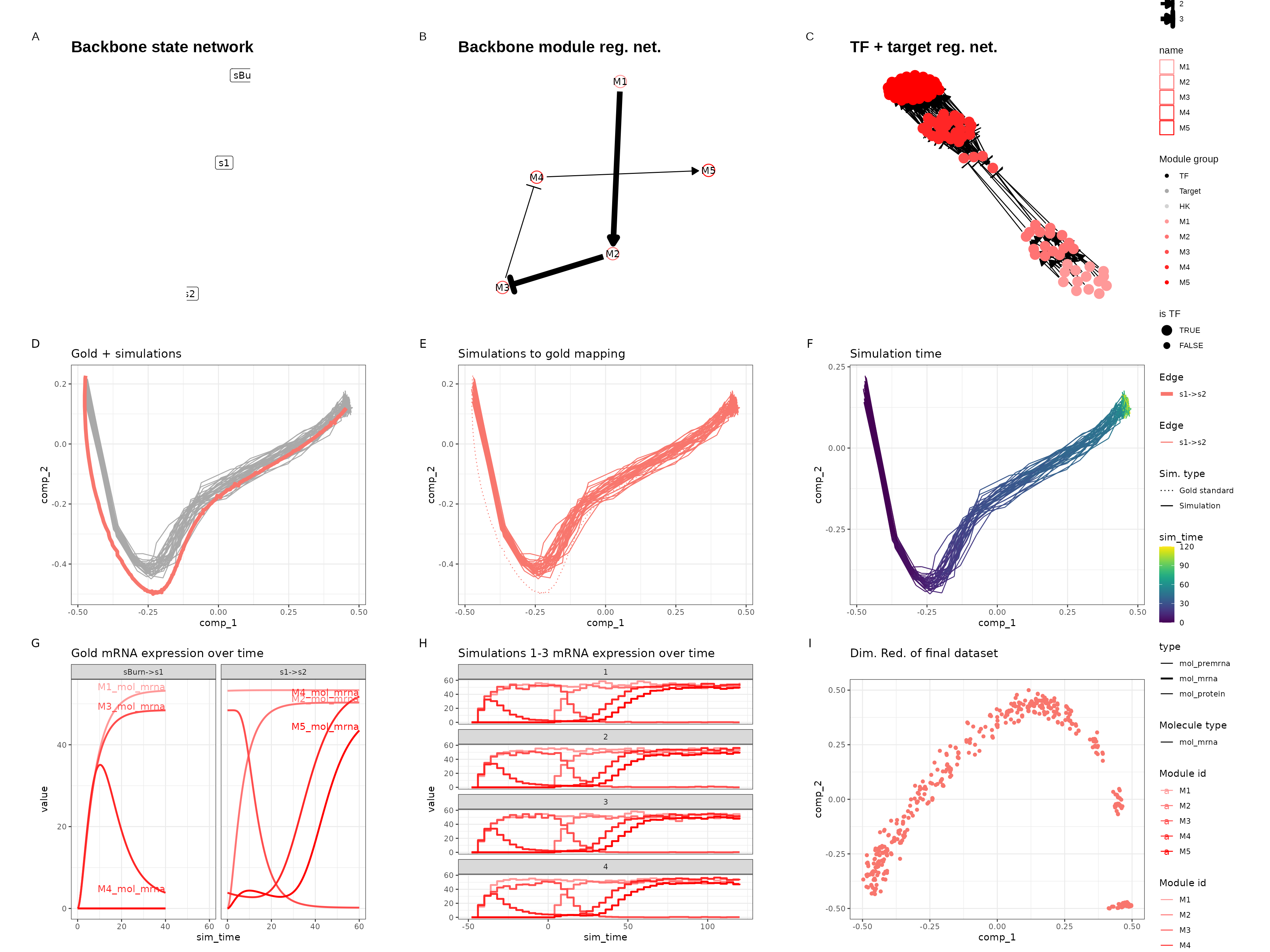
Cycle simple
set.seed(14)
backbone <- backbone_cycle_simple()
init <- initialise_model(
backbone = backbone,
num_cells = 500,
num_tfs = 100,
num_targets = 0,
num_hks = 0,
verbose = FALSE
)
out <- generate_dataset(init, make_plots = TRUE)
out$plot